Anaerobic microorganisms play important roles in human health and microbial green manufacturing. The gut microbiome, as the largest microbial community in the body, has evolved in conjunction with the host. The host provides a stable environment for microbial colonization, and the microorganisms, through their metabolism, participate in the digestion of complex dietary macromolecules and produce various compounds that have profound effects on host biology and play key roles in physiology. Research has shown that an imbalance in the gut microbiota is closely related to various human diseases. Most gut microorganisms are classified into the Bacteroidetes and Clostridia groups, which together make up more than 80% of the total microbial community. Although techniques such as 16S rRNA analysis and multi-omics methods have been widely used to study the association between microbial metabolites and host biology, a deeper understanding of the mechanisms behind the interaction between metabolites and the host/microbiota is still needed. To gain this knowledge, it is necessary to have relevant genetic manipulation tools for these symbiotic bacteria. Based on the known genome sequences of these two types of bacteria, researchers have developed genome editing tools for partial Bacteroides spp. and Clostridium spp., in order to better understand the interaction between these microorganisms and the host.
The Shewanella Biotechnology Research Group at the Qingdao Institute of Bioenergy and Bioprocess Technology recently published a review paper entitled "Advances in synthetic biology toolboxes paving the way for mechanistic understanding and strain engineering of gut commensal Bacteroides spp. and Clostridium spp." in the important biotechnology review journal Biotechnology Advances. The paper systematically summarized and reviewed the development and application of genome editing tools for gut commensal Bacteroides spp. and Clostridium spp., and Dr. Li Fuli is the corresponding author of this paper, while Dr. Tan Yang is the first author.
Regarding the aforementioned genome editing tools, constitutive and inducible promoters that can regulate gene expression have been developed. In addition, genetic knockout systems based on homologous recombination and transposons, as well as an exogenous DNA site-specific integration system based on phage integrases, have been established. Furthermore, genome editing tools based on CRISPR-Cas systems have also been developed, allowing for rapid and efficient editing of Bacteroides spp. and Clostridium spp. These synthetic biology tools enable us to gain deeper insights into the microbial-host and microbial-microbial interactions mediated by small molecule metabolites, and also allow us to manipulate the microbiome more rationally for the purpose of biotherapeutics. Based on this, the authors discuss future directions for microbiome editing tools and synthetic biology engineering research: (1) further improving the editing efficiency of new CRISPR-Cas-based genome editing tools, such as CRISPR-based base editing and transposon-related CRISPR-Cas systems; (2) developing in situ editing tools for microbial community editing as an alternative to traditional genetic methods that rely on manipulating individual species; (3) strain engineering of B. thetaiotaomicron and C. sporogenes is still in its early stages. Understanding their genomes is limited, and synthetic biology tools are needed to study the functions of their genome genes, expression regulation, interaction networks, and their impact on host biology. This knowledge is crucial for designing targeted effect molecules, sensors, and gene circuits.
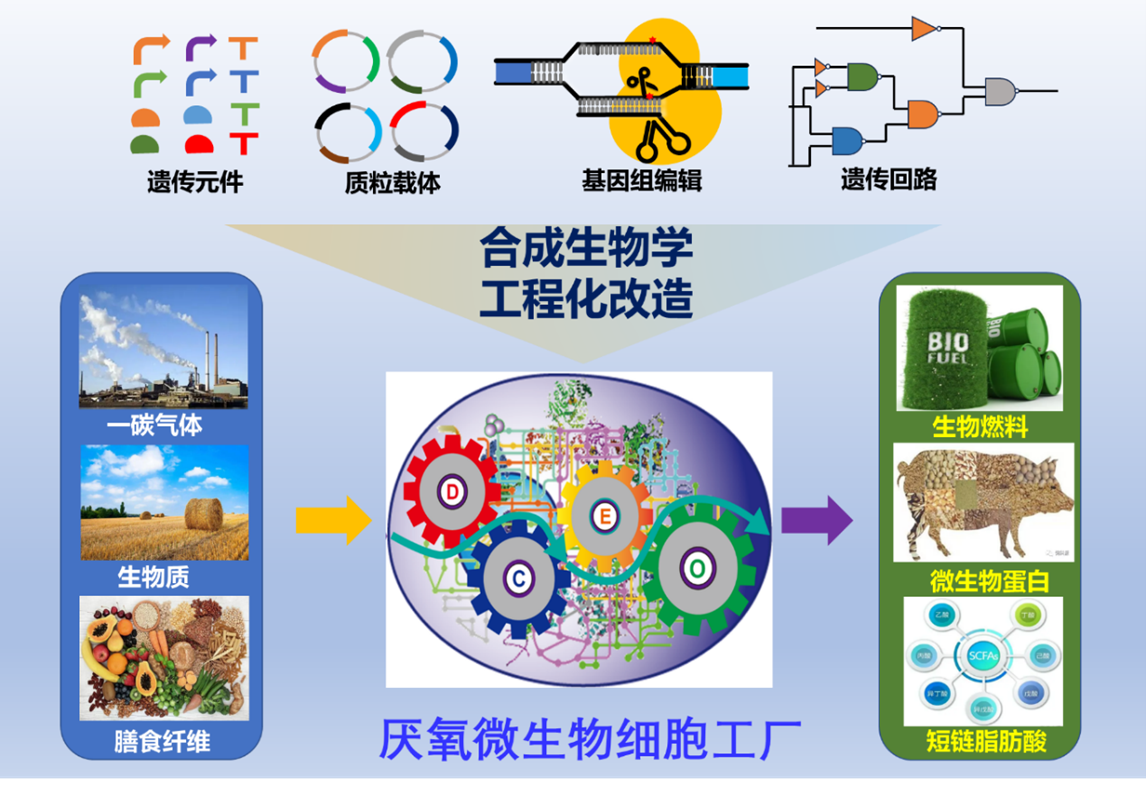
In addition, the Clostridium Biotechnology Research Group has published a review article titled "Recent progress in engineering Clostridium autoethanogenum to synthesize the biochemicals and biocommodities" in the journal "Synthetic and Systems Biotechnology." This review summarizes and discusses the research on metabolic engineering of C. autoethanogenum for the synthesis of chemicals and commodities using syngas. Dr. Tan Yang is the corresponding author, and Dr. Wan Sai is the first author of this article.
The transition utilization of fossil fuels has led to severe environmental consequences, exacerbating global warming and causing further climate change. In the face of these challenges, it is necessary to adopt multiple approaches to develop alternative solutions for sustainable chemicals and fuels. Microbial bio-catalytic synthesis processes have gained widespread attention in this context. For example, acetogenic bacteria can convert CO, CO2, and H2 into biomass and various metabolic products through the Wood-Ljungdahl pathway, which can be used for large-scale fermentation and the sustainable production of bulk biochemicals and biofuels such as acetic acid and ethanol. Clostridium autoethanogenum, as a representative acetogenic bacterium, serves as a model organism for microbial gas fermentation. In particular, the recent development of synthetic biology tools for this strain has allowed us to gain a deeper understanding of its unique physiological characteristics and achieve engineered transformations. This review summarizes research progress in the development of genome editing tools for C. autoethanogenum, the special energy metabolism models of autotrophic metabolism, and advances in metabolic engineering. As a platform with broad prospects for sustainable production of bulk chemicals, the engineering transformation of C. autoethanogenum still needs to address the following issues: (1) the development of high-throughput functional genomics tools to study gene interaction networks, providing new insights into the genotype-phenotype relationship; (2) the utilization of computer-assisted methods to overcome the metabolic engineering limitations imposed by the energy requirements of C. autoethanogenum in pathway design and enzyme engineering; (3) the further optimization of synthetic pathways through the development of synthetic biology component tools in recent years, including DNA genetic element libraries and the construction of genetic regulatory circuits to balance metabolic flux.
References:
This research is supported by the National Key R&D Program, the National Natural Science Foundation of China, the Chinese Academy of Sciences' Strategic Priority Research Program, Shandong Energy Research Institute, Qingdao New Energy Shandong Provincial Laboratory, and other projects. (Text/figure by Tan Yang)
Tan Y*, Liang J, Lai MC, Wan S, Luo XZ, Li FL*. 2023. Advances in synthetic biology toolboxes paving the way for mechanistic understanding and strain engineering of gut commensal Bacteroides spp. and Clostridium spp. Biotech Adv 69:108272 https://doi.org/10.1016/j.biotechadv.2023.108272.
Wan S, Lai MC, Gao XY, Zhou MX, Li FL, Xia L, Tan Y*. 2024. Recent progress in engineering Clostridium autoethanogenum to synthesize the biochemicals and biocommodities. Synth Syst Biotechnol, 9(1):19-25 https://doi.org/10.1016/j.synbio.2023.12.001.